Working with MPAS Grids#
Authors: Philip Chmielowiec, Ian Franda
This notebook showcases how to work with datasets from the Model for Prediction Across Scales (MPAS).
MPAS Grid Overview#
The defining feature of MPAS when compared to other models is that its unstructured grid is composed of Voronoi Meshes with a C-grid staggering. This means that the grid can be broken down into two meshes: Primal and Dual. The Primal Mesh is composed of Voronoi regions and the Dual Mesh is composed of Delaunay Triangles. The figure below showcases this relationship, with the dashed triangles being the dual of the Voronoi mesh.
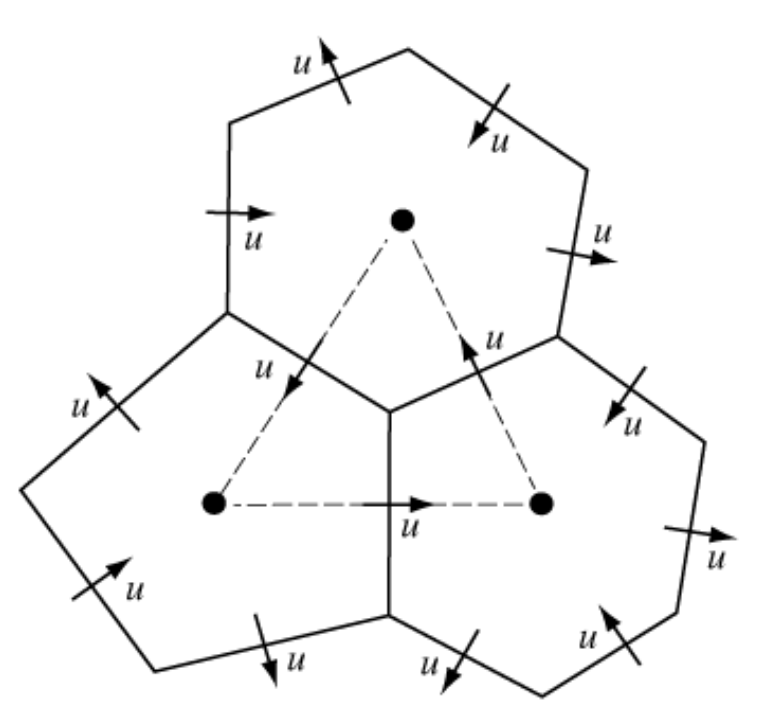
Since the Primal Mesh is predominantly made up of hexagons, with pentagons and heptagons occasionally being present, and the Dual Mesh being strictly triangular, this notebook will showcase how we can represent both of these meshes in the UGRID conventions using UXarray.
Dataset Overview#
Before diving straight into using UXarray, it is important to first investigate how MPAS datasets are represented.
As mentioned in earlier notebooks, the grid definition and data variables are typically stored in separate files. However, in this example, our dataset will contain both within the same file, which is often the case when working with smaller datasets. Additionally, even when working with separate Grid and Data files in MPAS, the definition of the Primal and Dual mesh are still stored under the same Grid file.
Below we can take a quick look into the dataset by opening it with Xarray.
# Dataset Path
mpas_root_filepath = "../../test/meshfiles/mpas/"
mpas_dataset_filepath = mpas_root_filepath + "QU/mesh.QU.1920km.151026.nc"
import xarray as xr
xrds_mpas = xr.open_dataset(mpas_dataset_filepath)
xrds_mpas
<xarray.Dataset> Size: 176kB
Dimensions: (nCells: 162, nEdges: 480, nVertices: 320,
maxEdges: 6, maxEdges2: 12, TWO: 2, vertexDegree: 3)
Dimensions without coordinates: nCells, nEdges, nVertices, maxEdges, maxEdges2,
TWO, vertexDegree
Data variables: (12/42)
latCell (nCells) float64 1kB ...
lonCell (nCells) float64 1kB ...
xCell (nCells) float64 1kB ...
yCell (nCells) float64 1kB ...
zCell (nCells) float64 1kB ...
indexToCellID (nCells) int32 648B ...
... ...
cellQuality (nCells) float64 1kB ...
gridSpacing (nCells) float64 1kB ...
triangleQuality (nVertices) float64 3kB ...
triangleAngleQuality (nVertices) float64 3kB ...
obtuseTriangle (nVertices) int32 1kB ...
meshDensity (nCells) float64 1kB ...
Attributes:
on_a_sphere: YES
sphere_radius: 1.0
is_periodic: NO
history: MpasMeshConverter.x base_grids/x1.162.grid.nc base_meshes...
mesh_spec: 1.0
Conventions: MPAS
source: MpasMeshConverter.x
file_id: rku96q0z66Here we opened up the dataset to get an overview of the full set of grid variables needed to describe an MPAS grid as outlined in the MPAS Specification Document [2]. Below is a list of the key grid variables that are used for representing and constructing the Primal and Dual meshes.
Primal Mesh#
lonVertex, latVertex: Corner Vertices of Primal Mesh cells
lonCell, latCell: Center Vertices of Primal Mesh cells
verticesOnCell: Vertex indices that surround each Primal Mesh cell
verticesOnEdge: Vertex indices that saddle a given edge
nEdgesOnCell: Maximum number of edges that can surround a cell
Dual Mesh#
lonCell, latCell: Corner Vertices of Dual Mesh cells
lonVertex, latVertex: Center Vertices of Dual Mesh cells
cellsOnVertex: Vertex indices that surround each Dual Mesh cell
cellsOnEdge: Vertex indices that saddle a given edge
Constructing a Grid Object#
Note
Since we only have a Grid file and no Data file in this example, we will be working exclusively with the Grid class to investigate the grid topology and not with UxDataset or UxDataArray data structures.
The xarray.Dataset that we opened in the previous section stores the coordinates and connectivity variables according to the MPAS specification standards for both the Primal and Dual meshes together in a single dataset. Here, instead of opening up the dataset using Xarray, we can pass through the path into our open_grid method to construct an instance of a Grid class. This Grid can take in a use_dual parameter to select whether to construct the Primal or Dual mesh, parsing and encoding the appropriate variables in the UGRID conventions.
import uxarray as ux
primal_mesh = ux.open_grid(mpas_dataset_filepath, use_dual=False)
dual_mesh = ux.open_grid(mpas_dataset_filepath, use_dual=True)
primal_mesh
<uxarray.Grid>
Original Grid Type: MPAS
Grid Dimensions:
* n_node: 320
* n_edge: 480
* n_face: 162
* n_max_face_nodes: 6
* n_max_face_edges: 6
* n_max_node_faces: 3
* two: 2
* n_nodes_per_face: (162,)
Grid Coordinates (Spherical):
* node_lon: (320,)
* node_lat: (320,)
* edge_lon: (480,)
* edge_lat: (480,)
* face_lon: (162,)
* face_lat: (162,)
Grid Coordinates (Cartesian):
* node_x: (320,)
* node_y: (320,)
* node_z: (320,)
* edge_x: (480,)
* edge_y: (480,)
* edge_z: (480,)
* face_x: (162,)
* face_y: (162,)
* face_z: (162,)
Grid Connectivity Variables:
* face_node_connectivity: (162, 6)
* face_edge_connectivity: (162, 6)
* edge_node_connectivity: (480, 2)
* edge_face_connectivity: (480, 2)
* node_face_connectivity: (320, 3)
Grid Descriptor Variables:
* edge_face_distances: (480,)
* edge_node_distances: (480,)
dual_mesh
<uxarray.Grid>
Original Grid Type: MPAS
Grid Dimensions:
* n_node: 162
* n_edge: 480
* n_face: 320
* n_max_face_nodes: 3
* n_max_face_edges: 3
* n_max_node_faces: 6
* two: 2
* n_nodes_per_face: (320,)
Grid Coordinates (Spherical):
* node_lon: (162,)
* node_lat: (162,)
* edge_lon: (480,)
* edge_lat: (480,)
* face_lon: (320,)
* face_lat: (320,)
Grid Coordinates (Cartesian):
* node_x: (162,)
* node_y: (162,)
* node_z: (162,)
* edge_x: (480,)
* edge_y: (480,)
* edge_z: (480,)
* face_x: (320,)
* face_y: (320,)
* face_z: (320,)
Grid Connectivity Variables:
* face_node_connectivity: (320, 3)
* face_edge_connectivity: (320, 3)
* edge_node_connectivity: (480, 2)
* edge_face_connectivity: (480, 2)
* node_face_connectivity: (162, 6)
Grid Descriptor Variables:
* edge_face_distances: (480,)
* edge_node_distances: (480,)
Relationship between MPAS and UGRID#
In the previous two sections, we outlined the set of grid variables used to describe the Primal and Dual meshes and how to open an MPAS grid in UXarray. Here, we provide an overview of how we represent both meshes in the UGRID conventions and how the original grid variables were modified to meet these conventions.
Grid Variables (Primal Mesh)#
node_lon & node_lat
Longitude and Latitude coordinates of the Primal Mesh corner nodes
Derived from
lonVertex&latVertexConverted from Radians to Degrees
face_lon & face_lat
Longitude and Latitude coordinates of the Primal Mesh center nodes
Derived from
lonCell&latCellConverted from Radians to Degrees
face_node_connectivity
Connectivity array describing which nodes make up a face
Derived from
verticesOnCellPadding is replaced with
INT_FILL_VALUEMissing Values (zeros) replaced with
INT_FILL_VALUEConverted to zero-index
edge_node_connectivity
Connectivity array describing which nodes link to form each edge
Derived from
verticesOnEdgePadding is replaced with
INT_FILL_VALUEMissing Values (zeros) replaced with
INT_FILL_VALUEConverted to zero-index
Grid Variables (Dual Mesh)#
node_lon & node_lat
Longitude and Latitude coordinates of the Dual Mesh vertices
Derived from
lonCell&latCell, the centers of the Primal MeshConverted from Radians to Degrees
face_lon & face_lat
Longitude and Latitude coordinates of the Dual Mesh centers
Derived from
lonVertex&latVertex, the vertices of the Primal MeshConverted from Radians to Degrees
face_node_connectivity
Connectivity array describing which nodes make up a face
Derived from
verticesOnCellPadding is replaced with
INT_FILL_VALUEMissing Values (zeros) replaced with
INT_FILL_VALUEConverted to zero-index
edge_node_connectivity
Connectivity array describing which nodes link to form each edge
Derived from
verticesOnEdgePadding is replaced with
INT_FILL_VALUEMissing Values (zeros) replaced with
INT_FILL_VALUEConverted to zero-index
Functionality#
Face Area Calculation#
Using our parsed attributes, we can determine whether our unstructured grid lies on the surface of a sphere by accessing the on_a_sphere attribute.
primal_mesh.parsed_attrs['on_a_sphere']
'YES'
Simiarly, we can access the sphere_radius attribute.
primal_mesh.parsed_attrs['sphere_radius']
1.0
Since our mesh lies on a sphere, we would expect our total surface area to follow the equation
\(4{\pi}{r^2}\)
We can use the value of the sphere_radius attribute to calculate the expected total surface area.
import numpy as np
sphere_r = primal_mesh.parsed_attrs['sphere_radius']
expected_area = 4 * np.pi * (sphere_r)**2
expected_area
12.566370614359172
UXarray can be used to compute the face area of each face on our grid.
primal_mesh_face_areas = primal_mesh.face_areas
primal_mesh_face_areas
<xarray.DataArray 'face_areas' (n_face: 162)> Size: 1kB
array([0.06733676, 0.06733676, 0.06733676, 0.06733676, 0.06733676,
0.06733676, 0.06733676, 0.06733676, 0.06733676, 0.06733676,
0.06733676, 0.06733676, 0.07879559, 0.07879561, 0.07879561,
0.07879559, 0.07879559, 0.07879559, 0.07879559, 0.07879559,
0.07879559, 0.07879561, 0.07879561, 0.07879559, 0.07879561,
0.07879561, 0.07879559, 0.07879559, 0.07879561, 0.07879559,
0.07879559, 0.07879561, 0.07879559, 0.07879559, 0.07879561,
0.07879559, 0.07879559, 0.07879559, 0.07879559, 0.07879559,
0.07879559, 0.07879561, 0.07631252, 0.07631253, 0.07631253,
0.07631252, 0.07631254, 0.07631254, 0.07631252, 0.07631253,
0.07631254, 0.07631252, 0.07631253, 0.07631254, 0.07631252,
0.07631254, 0.07631253, 0.07631254, 0.07631254, 0.07631253,
0.07631253, 0.07631252, 0.07631254, 0.07631253, 0.07631253,
0.07631252, 0.07631254, 0.07631253, 0.07631253, 0.07631252,
0.07631254, 0.07631253, 0.07631253, 0.07631253, 0.07631254,
0.07631253, 0.07631254, 0.07631253, 0.07631252, 0.07631252,
0.07631254, 0.07631253, 0.07631253, 0.07631252, 0.07631254,
0.07631254, 0.07631252, 0.07631254, 0.07631253, 0.07631253,
0.07631253, 0.07631253, 0.07631252, 0.07631254, 0.07631254,
0.07631252, 0.07631253, 0.07631252, 0.07631254, 0.07631254,
0.07631252, 0.07631254, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192, 0.08026192, 0.08026192, 0.08026192,
0.08026192, 0.08026192])
Dimensions without coordinates: n_face
Attributes:
cf_role: face_areasThe total face (surface) area can be computed by summing over each value.
primal_mesh_face_areas.sum()
<xarray.DataArray 'face_areas' ()> Size: 8B array(12.56637615)
We can then compute the absolute error of our calculation.
abs(expected_area - primal_mesh_face_areas.sum())
<xarray.DataArray 'face_areas' ()> Size: 8B array(5.53066503e-06)
The same can be done for the Dual Mesh.
dual_mesh_face_areas = dual_mesh.face_areas
dual_mesh_face_areas.sum()
<xarray.DataArray 'face_areas' ()> Size: 8B array(12.56637299)
abs(expected_area - dual_mesh_face_areas.sum())
<xarray.DataArray 'face_areas' ()> Size: 8B array(2.38060152e-06)
We can see that the total face area of both the Primal and Dual meshes is within 1e-6 of the expected area. For a more detailed explanation of the face area calculation and ways to obtain more precision, check out our other notebooks.
References#
[1] https://mpas-dev.github.io/
[2] https://mpas-dev.github.io/files/documents/MPAS-MeshSpec.pdf